Antibiotic resistance is a serious and growing phenomenon in contemporary medicine and has emerged as one of the pre-eminent public health concerns of the 21st century, particularly as it pertains to pathogenic organisms (the term is especially relevant to organisms which cause disease in humans). In the simplest cases, drug-resistant organisms may have acquired resistance to first-line antibiotics, thereby necessitating the use of second-line agents. Typically, a first-line agent is selected on the basis of several factors including safety, availability and cost; a second-line agent is usually broader in spectrum, has a less favourable risk-benefit profile and is more expensive or, in dire circumstances, be locally unavailable. In the case of some MDR pathogens, resistance to second and even third-line antibiotics is thus sequentially acquired, a case quintessentially illustrated by Staphylococcus aureus in some nosocomial settings. Some pathogens, such as Pseudomonas aeruginosa, also possess a high level of intrinsic resistance.
It may take the form of a spontaneous or induced genetic mutation, or the acquisition of resistance genes from other bacterial species by horizontal gene transfer via conjugation, transduction, or transformation. Many antibiotic resistance genes reside on transmissible plasmids, facilitating their transfer. Exposure to an antibiotic naturally selects for the survival of the organisms with the genes for resistance. In this way, a gene for antibiotic resistance may readily spread through an ecosystem of bacteria. Antibiotic-resistance plasmids frequently contain genes conferring resistance to several different antibiotics. This is not the case for Mycobacterium tuberculosis, the bacteria that causes Tuberculosis, since evidence is lacking for whether these bacteria have plasmids.[2] Also. M. tuberculosis lack the opportunity to interact with other bacteria in order to share plasmids.[2][3]
Genes for resistance to antibiotics, like the antibiotics themselves, are ancient.[4] However, the increasing prevalence of antibiotic-resistant bacterial infections seen in clinical practice stems from antibiotic use both within human medicine and veterinary medicine. Any use of antibiotics can increase selective pressure in a population of bacteria to allow the resistant bacteria to thrive and the susceptible bacteria to die off. As resistance towards antibiotics becomes more common, a greater need for alternative treatments arises. However, despite a push for new antibiotic therapies there has been a continued decline in the number of newly approved drugs.[5] Antibiotic resistance therefore poses a significant problem.
The growing prevalence and incidence of infections due to MDR pathogens is epitomised by the increasing number of familiar acronyms used to describe the causative agent and sometimes the infection generally; of these, MRSA is probably the most well-known, but others including VISA (vancomycin-intermediate S. aureus), VRSA (vancomycin-resistant S. aureus), ESBL (Extended spectrum beta-lactamase), VRE (Vancomycin-resistant Enterococcus) and MRAB (Multidrug-resistant A. baumannii) are prominent examples. Nosocomial infections overwhelmingly dominate cases where MDR pathogens are implicated, but multidrug-resistant infections are also becoming increasingly common in the community.
Contents
Causes
See also: Antibiotic misuse
Although there were low levels of preexisting antibiotic-resistant bacteria before the widespread use of antibiotics,[6] [7]
evolutionary pressure from their use has played a role in the
development of multidrug resistance varieties and the spread of
resistance between bacterial species.[8] In medicine, the major problem of the emergence of resistant bacteria is due to misuse and overuse of antibiotics.[9]
In some countries, antibiotics are sold over the counter without a
prescription, which also leads to the creation of resistant strains.
Other practices contributing towards resistance include antibiotic use in livestock feed.[10][11]
Household use of antibacterials in soaps and other products, although
not clearly contributing to resistance, is also discouraged (as not
being effective at infection control).[12]
Unsound practices in the pharmaceutical manufacturing industry can also
contribute towards the likelihood of creating antibiotic-resistant
strains.[13]
The procedures and clinical practice during the period of drug
treatment are frequently flawed — usually no steps are taken to isolate
the patient to prevent re-infection or infection by a new pathogen,
negating the goal of complete destruction by the end of the course.[14] (see Healthcare-associated infections and Infection control)Certain antibiotic classes are highly associated with colonisation with "superbugs" compared to other antibiotic classes. A superbug, also called multiresistant, is a bacterium that carries several resistance genes.[15] The risk for colonisation increases if there is a lack of susceptibility (resistance) of the superbugs to the antibiotic used and high tissue penetration, as well as broad-spectrum activity against "good bacteria". In the case of MRSA, increased rates of MRSA infections are seen with glycopeptides, cephalosporins and especially quinolones.[16][17] In the case of colonisation with Clostridium difficile the high risk antibiotics include cephalosporins and in particular quinolones and clindamycin.[18][19]
Of antibiotics used in the United States in 1997, half were used in humans and half in animals.[20]
Natural occurrence
There is evidence that naturally occurring antibiotic resistance is common.[21] The genes that confer this resistance are known as the environmental resistome.[21] These genes may be transferred from non-disease-causing bacteria to those that do cause disease, leading to clinically significant antibiotic resistance.[21]In 1952 an experiment conducted by Joshua and Esther Lederberg showed that penicillin-resistant bacteria existed before penicillin treatment.[22] While experimenting at the University of Wisconsin-Madison, Joshua Lederberg and his graduate student Norton Zinder also demonstrated preexistent bacterial resistance to streptomycin.[23] In 1962, the presence of penicillinase was detected in dormant Bacillus licheniformis endospores, revived from dried soil on the roots of plants, preserved since 1689 in the British Museum.[24][25][26] Six strains of Clostridium, found in the bowels of William Braine and John Hartnell (members of the Franklin Expedition) showed resistance to cefoxitin and clindamycin.[27] It was suggested that penicillinase may have emerged as a defense mechanism for bacteria in their habitats, such as the case of penicillinase-rich Staphylococcus aureus, living with penicillin-producing Trichophyton, however this was deemed circumstantial.[26] Search for a penicillinase ancestor has focused on the class of proteins that must be a priori capable of specific combination with penicillin.[28] The resistance to cefoxitin and clindamycin in turn was attributed to Braine's and Hartnell's contact with microorganisms that naturally produce them or random mutation in the chromosomes of Clostridium strains.[27] Nonetheless there is evidence that heavy metals and some pollutants may select for antibiotic-resistant bacteria, generating a constant source of them in small numbers.[29]
In medicine
The sheer volume of antibiotics prescribed is the major factor in the increasing rates of bacterial resistance rather than non-compliance with antibiotic protocol [30] A single dose of antibiotics leads to a greater risk of resistant organisms to that antibiotic in the person for up to a year.[31]Inappropriate prescribing of antibiotics has been attributed to a number of causes, including people who insist on antibiotics, physicians who simply prescribe them as they feel they do not have time to explain why they are not necessary, and physicians who do not know when to prescribe antibiotics or else are overly cautious for medical legal reasons.[32] For example, a third of people believe that antibiotics are effective for the common cold,[33] and the common cold is the most common reasons antibiotics are prescribed[34] even though antibiotics are completely useless against viruses.
Antibiotic resistance has been shown to increase with duration of treatment; therefore, as long as a clinically effective lower limit is observed (that depends upon the organism and antibiotic in question), the use by the medical community of shorter courses of antibiotics is likely to decrease rates of resistance, reduce cost, and have better outcomes due to fewer complications such as C. difficile infection and diarrhea.[35][36][37][38][39][40][41][42] In some situations a short course is inferior to a long course.[43] One study found that with one antibiotic a short course was more effective, but with a different antibiotic, a longer course was more effective.[44]
Advice to always complete a course of antibiotics is not based on strong evidence, and some researchers discourage the use of the prescription label “Finish all this medication unless otherwise directed by prescriber.” Often, antibiotics can be safely stopped 72 hours after symptoms resolve.[45] However, some infections require treatment long after symptoms are gone, and in all cases, an insufficient course of antibiotics may lead to relapse (with an infection that is now more antibiotic resistant).[46] Doctors must provide instructions to patients so they know when it is safe to stop taking a prescription since patients may feel better before the infection is eradicated. Some researchers advocate doctors' using a very short course of antibiotics, reevaluating the patient after a few days, and stopping treatment if there are no longer clinical signs of infection.[47]
A large number of people do not finish a course of antibiotics primarily because they feel better (varying from 10% to 44%, depending on the country).[48] Compliance with once-daily antibiotics is better than with twice-daily antibiotics.[49] Patients taking less than the required dosage or failing to take their doses within the prescribed timing results in decreased concentration of antibiotics in the bloodstream and tissues, and, in turn, exposure of bacteria to suboptimal antibiotic concentrations increases the frequency of antibiotic resistant organisms.[50]
Antibiotic-tolerant states may depend on physiological adaptations without direct connections to antibiotic target activity or to drug uptake, efflux, or inactivation. Identifying these adaptations, and targeting them to enhance the activity of existing drugs, is a promising approach to mitigate the public health crisis caused by the scarcity of new antibiotics.[citation needed]
Poor hand hygiene by hospital staff has been associated with the spread of resistant organisms,[51] and an increase in hand washing compliance results in decreased rates of these organisms.[52]
The improper use of antibiotics and therapeutic treatments can often be attributed to the presence of structural violence in particular regions. Socioeconomic factors such as race and poverty affect the accessibility of and adherence to drug therapy. The efficacy of treatment programs for these drug-resistant strains depends on whether or not programmatic improvements take into account the effects of structural violence.[53]
Role of other animals
Main article: Antibiotic use in livestock
Drugs are used in animals that are used as human food, such as
cattle, pigs, chickens, fish, etc. There has been extensive use of
antibiotics in animal husbandry.
Many of these drugs are not considered significant drugs for use in
humans, either because of their lack of efficacy or purpose in humans,
(such as the use of ionopores in ruminants[54]) or because that drug has gone out of use in humans (such as the decline in use of Sulfonamide_(medicine)
due to widespread allergic reactions and antibiotic resistance among
human pathogens.) Historically, regulation of antibiotic use in food
animals has been limited to limiting drug residues in meat, egg, and
milk products, rather than concern over the development of antibiotic
resistance. This mirrors the primary concerns in human medicine, where
researchers and doctors were historically more concerned about effective
but non-toxic doses of drugs rather than antibiotic resistance.
Evidence for the transfer of so-called superbugs from animals to humans
has been scant, and most evidence shows that pathogens of concern in
human populations originated in humans and are maintained there, with
rare cases of transference to humans. One of the pathogens most
frequently cited in popular literature — MRSA — is largely maintained in
the human population, often asyptomatically, and until recently has
rarely been found in food or companion animals.[55][56]
More significantly, evidence for the transference of fluoroquinolone
resistant genes in Campylobacteria strains through poultry was cited as
justification for severely restricting veterinary use of
fluoroquinolones in food animals in the USA. (Use in companion animals
is still permitted, and fluoroquinolones are the most commonly
prescribed antibiotic to adult humans in the United States, despite
guidelines which recommend it only be used in severe infections.)In 1945, in his Nobel Lecture, "Penillin," Alexander Fleming warned against the use of sub‐therapeutic doses of antibiotics – “bought by anyone in the shops” without a prescription: “The time may come when penicillin can be bought by anyone in the shops. Then there is the danger that the ignorant man may easily underdose himself and by exposing his microbes to non‐lethal quantities of the drug make them resistant. Here is a hypothetical illustration. Mr. X. has a sore throat. He buys some penicillin and gives himself, not enough to kill the streptococci but enough to educate them to resist penicillin. He then infects his wife. Mrs. X gets pneumonia and is treated with penicillin. As the streptococci are now resistant to penicillin the treatment fails. Mrs. X dies. Who is primarily responsible for Mrs. X’s death?”
In defiance of Fleming's teaching, since the late 1950s, massive quantities of non-prescription antibiotics, bought in farm fodder supplier "shops," have been fed in subtherapeutic doses to very large numbers of healthy livestock (pigs, beef cattle and poultry), for the sole purpose of increasing he weight by about six percent. All bacteria remaining alive in the manure of these animals must be resistant to the antibiotics used. Not only are they antibiotic resistant, but some of these "superbugs" acquire the ability to produce toxins, with some C. difficile organisms producing neurotoxins related to tetanus and botulinum toxins, two of the most powerful organic toxins known to man.
In 2001, the National Hog Farmer warned US producers that C. difficile “is sweeping the industry, killing many piglets” (Neutkens D; "New Clostridium Claiming Baby Pigs"). Thus, factory farms for antibiotic‐fed pigs are major reservoirs of C. difficile superbugs. Increasingly, C. difficile that has particularly afflicted hospitals in Quebec, a Canadian province with large numbers of pig factory farms.
"Superbugs," just by coexisting in the same environment (manure piles, contaminated water, in the throats or colons of infected patients, or hospital staff carriers) have the ability to share antibiotic resistant genesby "gene jumping," a mechanism unknown by and unimaginable to Fleming (Google: "antibiotic resistance, gene jumping").
The resistant bacteria in animals due to antibiotic exposure can be transmitted to humans via three pathways, those being through the consumption of meat, from close or direct contact with animals, or through the environment.[57]/ However, complete cooking of meat inactivates bacteria, whether or not they are antibiotic-resistant. The World Health Organization concluded antibiotics as growth promoters in animal feeds should be prohibited, in the absence of risk assessments.
In 1998, European Union health ministers voted to ban four antibiotics widely used to promote animal growth (despite their scientific panel's recommendations). Regulation banning the use of antibiotics in European feed, with the exception of two antibiotics in poultry feeds, became effective in 2006.[58] In Scandinavia, there is evidence that the ban has led to a lower prevalence of antimicrobial resistance in (nonhazardous) animal bacterial populations.[59]
In the United States
In the USA, federal agencies do not collect data on antibiotic use in animals, but animal-to-human spread of drug-resistant organisms has been demonstrated in research studies. Antibiotics are still used in U.S. animal feed, along with other ingredients that represent safety concerns.[11][60]Growing U.S. consumer concern about using antibiotics in animal feed has led to a niche market of "antibiotic-free" animal products, but this small market is unlikely to change entrenched, industry-wide practices.[61]
In 2001, the Union of Concerned Scientists estimated that greater than 70% of the antibiotics used in the US are given to food animals (for example, chickens, pigs and cattle), in the absence of disease.[62] Hence, the amounts given are termed "sub-therapeutic", i.e., insufficient to combat disease—because no demonstrable disease is present. Sub-therapeutic dosages kills some, but not all, of the bacterial organisms in the animal—leaving those that are naturally antibiotic-resistant. Studies have shown, however, that the overall population levels of bacteria are essentially unchanged; only the mix of bacteria is affected.[citation needed]
Thus the actual mechanism by which sub-therapeutic antibiotic feed additives serve as growth promotors is unclear. Some people have speculated that animals and fowl in feedlot environments may have sub-clinical infections, which are cured by low levels of antibiotics in feed, thereby allowing the creatures to thrive; but no convincing evidence has been advanced for this theory. As the bacterial load in an animal is essentially unchanged by use of antibiotic feed additives, the mechanism of growth promotion is overwhelmingly likely to be something other than "killing off the bad bugs."
In 2000, the US Food and Drug Administration (FDA) announced their intention to revoke approval of fluoroquinolone use in poultry production because of substantial evidence linking it to the emergence of fluoroquinolone-resistant Campylobacter infections in humans. The final decision to ban fluoroquinolones from use in poultry production was not made until five years later because of challenges from the food animal and pharmaceutical industries.[63]
During 2007, two federal bills (S. 549[64] and H.R. 962[65]) aimed at phasing out "nontherapeutic" antibiotics in US food animal production. The Senate bill, introduced by Sen. Edward "Ted" Kennedy, died. The House bill, introduced by Rep. Louise Slaughter, died after being referred to Committee.
In the United States, evidence of emergence of antibiotic-resistant bacterial strains due to wide use of antibiotics to promote weight gain in livestock was determined by the United States Food and Drug Administration in 1977, but nothing effective was done to prevent the practice. In March, 2012 the United States District Court for the Southern District of New York, ruling in an action brought by the Natural Resources Defense Council and others, ordered the FDA to revoke approvals for the use of antibiotics in livestock which violated FDA regulations.[66] On April 11, 2012 the FDA announced a voluntary program to phase out unsupervised use of drugs as feed additives and convert approved over-the-counter uses for antibiotics to prescription use only, requiring veterinarian supervision of their use and a prescription.[67][68]
Environmental Impact
Antibiotics have been polluting the environment since their introduction through human waste (medication, farming), animals, and the pharmaceutical industry.[69] Along with antibiotic waste, resistant bacteria follow, thus introducing antibiotic resistant bacteria into the environment. As bacteria replicate quickly, the resistant bacteria that enter the environment replicate their resistance genes as they continue to divide. Additionally, bacteria carrying resistance genes have the ability to spread those genes to other species via horizontal gene transfer. Therefore, even if the specific antibiotic is no longer introduced into the environment, antibiotic resistance genes will persist through the bacteria that have since replicated without continual exposure.[69]A study done of the Poudre River implicated wastewater treatment plants, as well as animal feeding operations in the dispersal of antibiotic resistance genes into the environment.[70] This research was done using molecular signatures in order to determine the sources, and the location at the Poudre River was chosen due to lack of other anthropogenic influences upstream. The study indicates that monitoring of antibiotic resistance genes may be useful in determining not only the point of origin of their release, but also how these genes persist in the environment. Additionally, studying physical and chemical methods of treatment may alleviate pressure of antibiotic resistance genes in the environment, and thus their entry back into human contact.
Mechanisms
See also: Microevolution

Schematic representation of how antibiotic resistance evolves via
natural selection. The top section represents a population of bacteria
before exposure to an antibiotic. The middle section shows the
population directly after exposure, the phase in which selection took
place. The last section shows the distribution of resistance in a new
generation of bacteria. The legend indicates the resistance levels of
individuals.
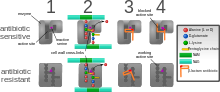
Diagram depicting antibiotic resistance through alteration of the
antibiotic's target site, modeled after MRSA's resistance to penicillin.
Beta-lactam antibiotics permanently inactivate PBP enzymes, which are essential for bacterial life, by permanently binding to their active sites. MRSA, however, expresses a PBP that does not allow the antibiotic into its active site.
The four main mechanisms by which microorganisms exhibit resistance to antimicrobials are:
- Drug inactivation or modification: for example, enzymatic deactivation of penicillin G in some penicillin-resistant bacteria through the production of β-lactamases
- Alteration of target site: for example, alteration of PBP—the binding target site of penicillins—in MRSA and other penicillin-resistant bacteria
- Alteration of metabolic pathway: for example, some sulfonamide-resistant bacteria do not require para-aminobenzoic acid (PABA), an important precursor for the synthesis of folic acid and nucleic acids in bacteria inhibited by sulfonamides, instead, like mammalian cells, they turn to using preformed folic acid.
- Reduced drug accumulation: by decreasing drug permeability and/or increasing active efflux (pumping out) of the drugs across the cell surface[72]
Antibiotic resistance can also be introduced artificially into a microorganism through laboratory protocols, sometimes used as a selectable marker to examine the mechanisms of gene transfer or to identify individuals that absorbed a piece of DNA that included the resistance gene and another gene of interest. A recent study demonstrated that the extent of horizontal gene transfer among Staphylococcus is much greater than previously expected—and encompasses genes with functions beyond antibiotic resistance and virulence, and beyond genes residing within the mobile genetic elements.[76]
For a long time it has been thought that for a microorganism to become resistant to an antibiotic, it must be in a large population. However, recent findings show that there is no necessity of large populations of bacteria for the appearance of antibiotic resistance. We know now, that small populations of E.coli in an antibiotic gradient can become resistant. Any heterogeneous environment with respect to nutrient and antibiotic gradients may facilitate the development of antibiotic resistance in small bacterial populations and this is also true for the human body. Researchers hypothesize that the mechanism of resistance development is based on four SNP mutations in the genome of E.coli produced by the gradient of antibiotic. These mutations confer the bacteria emergence of antibiotic resistance.
A common misconception is that a person can become resistant to certain antibiotics. It is a strain of microorganism that can become resistant, not a person's body.[77][78]
Resistant pathogens
Staphylococcus aureus
Main article: Methicillin-resistant Staphylococcus aureus
Staphylococcus aureus (colloquially known as "Staph aureus" or a "Staph infection") is one of the major resistant pathogens. Found on the mucous membranes and the human skin
of around a third of the population, it is extremely adaptable to
antibiotic pressure. It was one of the earlier bacteria in which penicillin resistance was found—in 1947, just four years after the drug started being mass-produced. Methicillin was then the antibiotic of choice, but has since been replaced by oxacillin due to significant kidney toxicity. Methicillin-resistant Staphylococcus aureus
(MRSA) was first detected in Britain in 1961, and is now "quite common"
in hospitals. MRSA was responsible for 37% of fatal cases of sepsis in the UK in 1999, up from 4% in 1991. Half of all S. aureus infections in the US are resistant to penicillin, methicillin, tetracycline and erythromycin.This left vancomycin as the only effective agent available at the time. However, strains with intermediate (4-8 μg/ml) levels of resistance, termed glycopeptide-intermediate Staphylococcus aureus (GISA) or vancomycin-intermediate Staphylococcus aureus (VISA), began appearing in the late 1990s. The first identified case was in Japan in 1996, and strains have since been found in hospitals in England, France and the US. The first documented strain with complete (>16 μg/ml) resistance to vancomycin, termed vancomycin-resistant Staphylococcus aureus (VRSA) appeared in the United States in 2002.[79] However, in 2011 a variant of vancomycin has been tested that binds to the lactate variation and also binds well to the original target, thus reinstates potent antimicrobial activity.[80]
A new class of antibiotics, oxazolidinones, became available in the 1990s, and the first commercially available oxazolidinone, linezolid, is comparable to vancomycin in effectiveness against MRSA. Linezolid-resistance in S. aureus was reported in 2001.[81]
Community-acquired MRSA (CA-MRSA) has now emerged as an epidemic that is responsible for rapidly progressive, fatal diseases, including necrotizing pneumonia, severe sepsis and necrotizing fasciitis.[82] MRSA is the most frequently identified antimicrobial drug-resistant pathogen in US hospitals. The epidemiology of infections caused by MRSA is rapidly changing. In the past 10 years[when?], infections caused by this organism have emerged in the community. The two MRSA clones in the United States most closely associated with community outbreaks, USA400 (MW2 strain, ST1 lineage) and USA300, often contain Panton-Valentine leukocidin (PVL) genes and, more frequently, have been associated with skin and soft tissue infections. Outbreaks of CA-MRSA infections have been reported in correctional facilities, among athletic teams, among military recruits, in newborn nurseries, and among men who have sex with men. CA-MRSA infections now appear endemic in many urban regions and cause most CA-S. aureus infections.[83]
Streptococcus and Enterococcus
Streptococcus pyogenes (Group A Streptococcus: GAS) infections can usually be treated with many different antibiotics. Early treatment may reduce the risk of death from invasive group A streptococcal disease. However, even the best medical care does not prevent death in every case. For those with very severe illness, supportive care in an intensive care unit may be needed. For persons with necrotizing fasciitis, surgery often is needed to remove damaged tissue.[84] Strains of S. pyogenes resistant to macrolide antibiotics have emerged; however, all strains remain uniformly susceptible to penicillin.[85]Resistance of Streptococcus pneumoniae to penicillin and other beta-lactams is increasing worldwide. The major mechanism of resistance involves the introduction of mutations in genes encoding penicillin-binding proteins. Selective pressure is thought to play an important role, and use of beta-lactam antibiotics has been implicated as a risk factor for infection and colonization. S. pneumoniae is responsible for pneumonia, bacteremia, otitis media, meningitis, sinusitis, peritonitis and arthritis.[85]
Multidrug-resistant Enterococcus faecalis and Enterococcus faecium are associated with nosocomial infections.[86] Among these strains, penicillin-resistant Enterococcus was seen in 1983, vancomycin-resistant Enterococcus in 1987, and linezolid-resistant Enterococcus in the late 1990s.[citation needed]
Pseudomonas aeruginosa
Pseudomonas aeruginosa is a highly prevalent opportunistic pathogen. One of the most worrisome characteristics of P. aeruginosa is its low antibiotic susceptibility, which is attributable to a concerted action of multidrug efflux pumps with chromosomally encoded antibiotic resistance genes (for example, mexAB-oprM, mexXY, etc.) and the low permeability of the bacterial cellular envelopes.[87] Pseudomonas aeruginosa has the ability to produce HAQs and it has been found that HAQs have prooxidant effects, and overexpressing modestly increased susceptibility to antibiotics. The study experimented with the Pseudomonas aeruginosa biofilms and found that a disruption of relA and spoT genes produced an inactivation of the Stringent response (SR) in cells who were with nutrient limitation which provides cells be more susceptible to antibiotics.[88]Clostridium difficile
Clostridium difficile is a nosocomial pathogen that causes diarrheal disease in hospitals world wide.[89][90] Clindamycin-resistant C. difficile was reported as the causative agent of large outbreaks of diarrheal disease in hospitals in New York, Arizona, Florida and Massachusetts between 1989 and 1992.[91] Geographically dispersed outbreaks of C. difficile strains resistant to fluoroquinolone antibiotics, such as ciprofloxacin and levofloxacin, were also reported in North America in 2005.[92]Salmonella and E. coli
Infection with Escherichia coli and Salmonella can result from the consumption of contaminated food. When both bacteria are spread, serious health conditions arise. Many people are hospitalized each year after becoming infected, with some dying as a result. Since 1993, some strains of E. coli have become resistant to multiple types of fluoroquinolone antibiotics.[citation needed]Acinetobacter baumannii
On November 5, 2004, the Centers for Disease Control and Prevention (CDC) reported an increasing number of Acinetobacter baumannii bloodstream infections in patients at military medical facilities in which service members injured in the Iraq/Kuwait region during Operation Iraqi Freedom and in Afghanistan during Operation Enduring Freedom were treated. Most of these showed multidrug resistance (MRAB), with a few isolates resistant to all drugs tested.[93][94]Mycobacterium tuberculosis
Tuberculosis is increasing across the globe, especially in developing countries, over the past few years. TB resistant to antibiotics is called MDR TB (Multidrug Resistant TB). The rise of the HIV/AIDS epidemic has contributed to this.[95]TB was considered one of the most prevalent diseases, and did not have a cure until the discovery of Streptomycin by Selman Waksman in 1943.[96] However, the bacteria soon developed resistance. Since then, drugs such as isoniazid and rifampin have been used. M. tuberculosis develops resistance to drugs by spontaneous mutations in its genomes. Resistance to one drug is common, and this is why treatment is usually done with more than one drug. Extensively Drug Resistant TB (XDR TB) is TB that is also resistant to the second line of drugs.[95][97]
Resistance of Mycobacterium tuberculosis to isoniazid, rifampin, and other common treatments has become an increasingly relevant clinical challenge. (For more on Drug Resistant TB, visit the Multi-drug resistant tuberculosis page.)
Alternatives
Prevention
Rational use of antibiotics may reduce the chances of development of opportunistic infection by antibiotic-resistant bacteria due to dysbacteriosis.Our immune systems will cure minor bacterial infections on their own. If we give it the chance without relying on antibiotics to cure a small infection, we will be less likely to become immune or resistant to the antibiotic. It is also important to note that antibiotics will not cure viral infections. Taking an antibiotic unnecessarily to treat a viral infection can lead to the resistance of antibiotics[98] In one study, the use of fluoroquinolones is clearly associated with Clostridium difficile infection, which is a leading cause of nosocomial diarrhea in the United States,[99] and a major cause of death, worldwide.[100]Vaccines do not have the problem of resistance because a vaccine enhances the body's natural defenses, while an antibiotic operates separately from the body's normal defenses. Nevertheless, new strains may evolve that escape immunity induced by vaccines; for example an updated influenza vaccine is needed each year.
While theoretically promising, antistaphylococcal vaccines have shown limited efficacy, because of immunological variation between Staphylococcus species, and the limited duration of effectiveness of the antibodies produced. Development and testing of more effective vaccines is under way.[101]
The Australian Commonwealth Scientific and Industrial Research Organization (CSIRO), realizing the need for the reduction of antibiotic use, has been working on two alternatives. One alternative is to prevent diseases by adding cytokines instead of antibiotics to animal feed.[102] These proteins are made in the animal body "naturally" after a disease and are not antibiotics, so they do not contribute to the antibiotic resistance problem. Furthermore, studies on using cytokines have shown they also enhance the growth of animals like the antibiotics now used, but without the drawbacks of nontherapeutic antibiotic use. Cytokines have the potential to achieve the animal growth rates traditionally sought by the use of antibiotics without the contribution of antibiotic resistance associated with the widespread nontherapeutic uses of antibiotics currently used in the food animal production industries. Additionally, CSIRO is working on vaccines for diseases.[102]
Phage therapy
Phage therapy, an approach that has been extensively researched and used as a therapeutic agent for over 60 years, especially in the Soviet Union, represents a potentially significant but currently underdeveloped approach to the treatment of bacterial disease.[103] Phage therapy was widely used in the United States until the discovery of antibiotics, in the early 1940s. Bacteriophages or "phages" are viruses that invade bacterial cells and, in the case of lytic phages, disrupt bacterial metabolism and cause the bacterium to lyse. Phage therapy is the therapeutic use of lytic bacteriophages to treat pathogenic bacterial infections.[104] [105] [106]Bacteriophage therapy is an important alternative to antibiotics in the current era of multidrug resistant pathogens. A review of studies that dealt with the therapeutic use of phages from 1966–1996 and few latest ongoing phage therapy projects via internet showed: phages were used topically, orally or systemically in Polish and Soviet studies. The success rate found in these studies was 80–95% with few gastrointestinal or allergic side effects. British studies also demonstrated significant efficacy of phages against Escherichia coli, Acinetobacter spp., Pseudomonas spp. and Staphylococcus aureus. US studies dealt with improving the bioavailability of phage. Phage therapy may prove as an important alternative to antibiotics for treating multidrug resistant pathogens.[107]
Discovery of the structure of the viral protein PlyC is allowing researchers to understand the way it kills a significant range of pathogenic bacteria.[108]
Research
Development pipeline
Since the discovery of antibiotics, research and development (R&D) efforts have provided new drugs in time to treat bacteria that became resistant to older antibiotics, but in the 2000s there has been concern that development has slowed enough that seriously ill patients may run out of treatment options.[109] Another concern is that doctors may become reluctant to perform routine surgeries due to the increased risk of harmful infection.[110] Backup treatments can have serious side effects; for example, treatment of multi drug resistant tuberculosis can cause deafness and insanity.[111] The potential crisis at hand is the result of a marked decrease in industry R&D.[112] Poor financial investment in antibiotic research has exacerbated the situation.[112]In the United States, drug companies and the administration of President Barack Obama have been proposing changing the standards by which the FDA approves antibiotics targeted at resistant organisms.[113][110] The U.S. National Institutes of Health plans to fund a new research network on the issue up to $62 million from 2013 to 2019.[114] Using authority created by the Pandemic and All Hazards Preparedness Act of 2006, the Biomedical Advanced Research and Development Authority in the U.S. Department of Health and Human Services announced it will spend between $40 million and $200 million in funding for R&D on new antibiotic drugs under development by GlaxoSmithKline.[115]
Mechanism
In research published on October 17, 2008 in Cell, a team of scientists pinpointed the place on bacteria where the antibiotic myxopyronin launches its attack, and why that attack is successful. The myxopyronin binds to and inhibits the crucial bacterial enzyme, RNA polymerase. The myxopyronin changes the structure of the switch-2 segment of the enzyme, inhibiting its function of reading and transmitting DNA code. This prevents RNA polymerase from delivering genetic information to the ribosomes, causing the bacteria to die.[116]In 2012, a team of the University of Leipzig modified a peptide found in honeybees. It is effective against 37 types of bacteria.[117]
One major cause of antibiotic resistance is the increased pumping activity of microbial ABC transporters, which diminishes the effective drug concentration inside the microbial cell. ABC transporter inhibitors that can be used in combination with current antimicrobials are being tested in clinical trials and are available for therapeutic regimens.[118]
Applications
Antibiotic resistance is an important tool for genetic engineering. By constructing a plasmid that contains an antibiotic resistance gene as well as the gene being engineered or expressed, a researcher can ensure that when bacteria replicate, only the copies that carry the plasmid survive. This ensures that the gene being manipulated passes along when the bacteria replicates.The most commonly used antibiotics in genetic engineering are generally "older" antibiotics that have largely fallen out of use in clinical practice. These include:
Industrially the use of antibiotic resistance is disfavored since maintaining bacterial cultures would require feeding them large quantities of antibiotics. Instead, the use of auxotrophic bacterial strains (and function-replacement plasmids) is preferred.
See also
- Alliance for the Prudent Use of Antibiotics
- Antibacterial soap
- Beta-lactamase#KPC (K. pneumoniae carbapenemase) (Class A) (KPC) antibacterial resistance gene
- Broad-spectrum antibiotic
- Carbapenem resistant enterobacteriaceae
- Center for Disease Dynamics, Economics & Policy
- Drug of last resort
- Index of environmental articles
- Multidrug tolerance
- Multidrug-resistant gram-negative bacteria
- New Delhi metallo-beta-lactamase (NDM-1) antibacterial resistance gene
- Pesticide resistance


No comments:
Post a Comment